A MATLAB Based Toolbox for Processing and Analysis of Multichannel, Multisource Cortical Local Field Potentials
Mufti Mahmud (NeuroChip Lab, University of Padova), Alessandra Bertoldo (Dept. of Information Engineering, University of Padova), Stefano Girardi (NeuroChip Lab, University of Padova), Marta Maschietto (NeuroChip Lab, University of Padova), Stefano Vassanelli (NeuroChip Lab, University of Padova)
The neuronal probe technology has experienced a rapid development during the last decade and enabled the scientists to record neuronal signals from multiple sites / regions simultaneously generating an enormous amount of data. To infer meaningful conclusions by processing and analyzing this massive amount of data have been a big challenge to the neuroscience and neuroengineering community [1], [2]. To respond to this challenge some research groups have developed their own automated processing and analysis tools; also, some are available commercially. However, almost all of these tools address single spikes [3], [4] while the local field potentials (LFPs) have been left unaddressed. Therefore, to bridge this gap, we have developed a Matlab based toolbox, named “SigMate”, capable of performing processing and analysis of cortical LFPs.
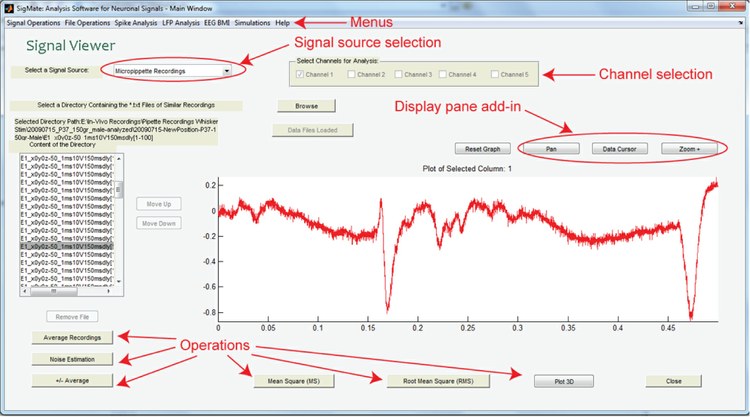
The main aim behind the development of SigMate is to deliver an opensource tool to the community which can be easily modified and adapted to LFPs recorded from different experimental conditions and different cortical regions. The modular approach in SigMate’s design will allow it to serve as a framework for the developers in the community. It can process and analyze signals from multisite neuronal probes as well as multiple sources (conventional borosilicate micropipettes, and multichannel state-of-the-art neuronal probe) facilitating comparative studies. The features of SigMate are growing with time, however, available features at present are: data visualization (2D and 3D), file operations (file splitting, concatenation, and column rearranging), slow stimulus artifact removal (including baseline correction), fast stimulus artifact removal from LFPs induced by intracortical microstimulation, noise characterization, latency estimation, current source density (CSD) analysis, determination of cortical layer activation order (CLAO) from LFPs and CSDs, single sweep LFP classification, and are growing. The figure 1 shows the initial graphical user interface (GUI) of the SigMate containing the data visualization feature and the menu bar for accessing the other features.
The available features of SigMate (i.e., our in-house algorithms) have been extensively tested using LFPs recorded from anesthetized rats by standard borosilicate micropipettes (1 MΩ resistance) and implantable Electrolyte-Oxide-Semiconductor Field Effect Transistor (EOSFET) based multisite neural probes. It will soon be made available to the community under the GNU-GPL [5].
References
- G. Buzsaki, Large-scale recording of neuronal ensembles. Nat. Neurosci. 7(5): 446-451, 2004.
- K.D. Wise, D.J. Anderson, J.F. Hetke, D.R. Kipke, K. Najafi, Wireless Implantable Microsystems: High-Density Electronic Interfaces to the Nervous System. Proc. of the IEEE 92(1): 76-97, 2004.
- L. Hazan, M. Zugaro, G. Buzsaki, Klusters, NeuroScope, NDManager: A free software suite for neurophysiological data processing and visualization. J. Neurosci. Meth. 155: 207–216, 2006.
- M. Lidierth, sigTOOL: A MATLAB-based environment for sharing laboratory-developed software to analyze biological signals. J. Neurosci. Meth. 178: 188–196, 2009.
- M. Mahmud, A. Bertoldo, S. Girardi, M. Maschietto, S. Vassanelli, SigMate: A MATLAB-based Neuronal Signal Processing Tool. In: Proc. of the 32nd IEEE EMBC2010, Buenos Aires, September 2010, pp. 1352-1355.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
