A neuroinformatics framework for gene expression meta-analysis
Paul Pavlidis (University of British Columbia), Anton Zoubarev (University of British Columbia), Kelsey Hamer (University of British Columbia), Kiran Keshav (Columbia University)
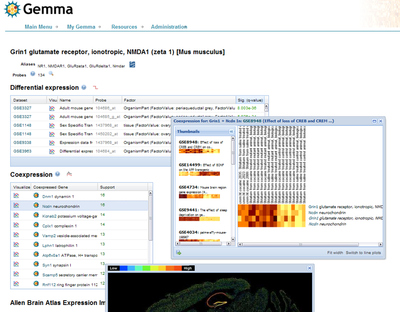
We describe the development and application of “Gemma” (www.chibi.ubc.ca/Gemma), a neuroinformatics system for the large-scale analysis of genomics data, focused on expression meta-analysis. The database contains over 2300 expression studies (“data sets”). Over 700 of these are brain-related. In total there are over 87,000 microarray assays in the system. Data sets are analyzed and annotated using automated tools (enhanced by extensive manual curation) using the NIFStd Ontology and other ontologies. Each data set is quality-controlled and analyzed for expression patterns of interest in two ways. First, we use “coexpression”, which reflect correlations of expression among genes across conditions. This can be viewed as inferring gene networks which in turn can inform functional analyses. Second, data sets are analyzed for differential expression, which are changes in expression with respect to experimental covariates such as genotype or drug treatment. The system contains well over a billion data points from these analyses.
The Gemma web interface allows the browsing, searching, download and visualization of these data and the analysis results. We also offer web services and a Cytoscape plugin for viewing coexpression networks. Gemma is designed as a data sharing tool. Users can upload, annotate and analyze their own expression profiling data. Gemma implements a security model which includes the concepts of public, shared or private data, and “user groups”. Users can also create and share gene groups and dataset groups as well as uploading their own data. This allows users to compare their data sets with other data sets including the hundreds of public data sets. Gemma also has the concept of “gene groups” and “dataset groups”, which can be defined by us or by registered users. Thus Gemma can be used as a collaborative tool, and to analyze expression data sets prior to making them public. The database also includes information on protein interactions, rodent brain expression patterns from the the Allen Brain Atlas, transcriptional regulation from Pazar, and contains additional information obtained from analyses we have performed of specific relevance to neuroscientists. We will describe ongoing efforts to integrate data from Gemma with other neuroinformatics resources including the Neuroscience Information Framework (NIF) and GeneNetwork.org.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
