BrainTrap Comparisons: Fly Brain Protein Trap Data Analysis
Seymour Knowles-Barley (The University of Edinburgh), J Douglas Armstrong (The University of Edinburgh)
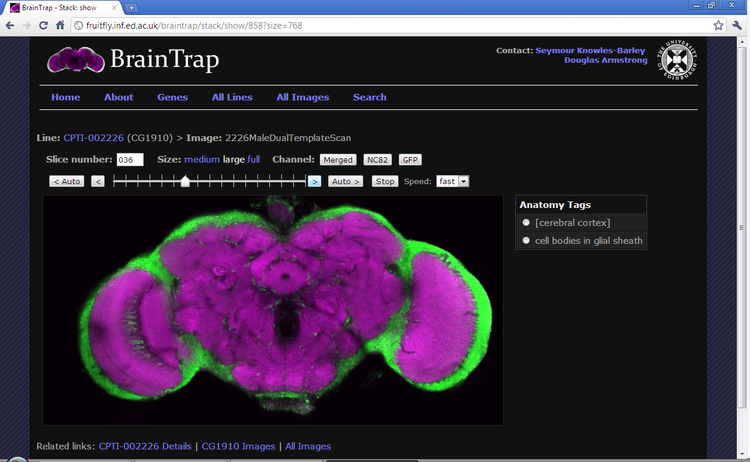
Protein-trap strains of Drosophila melanogaster provide a very useful tool for examining the 3D-expression patterns of proteins and purification of protein complexes. BrainTrap, the fly brain protein trap database, contains 3D confocal images of over 500 Drosophila protein trap lines. Full size images can be viewed and annotated interactively in a web browser. The database includes searchable annotations highlighting the anatomical location of expression linked to the FlyBase anatomical ontology.
The BrainTrap database is available online at:
http://fruitfly.inf.ed.ac.uk/braintrap
In addition we have compared the anatomical areas of expression to gene expression data in the mouse brain available in the Allen Brain Atlas. We find that general expression in fruitfly neuropil areas correlates with higher levels of expression in the mouse and targeted expression in some areas of the fly brain correlates with higher expression in a subset of areas in the mouse.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
