Constrain your data: relating detailed animal studies to spatial templates in neuroimaging
Gleb Bezgin (Rotman Research Institute at Baycrest ), Rembrandt Bakker (Donders Institute for Brain, Cognition and Behavior)
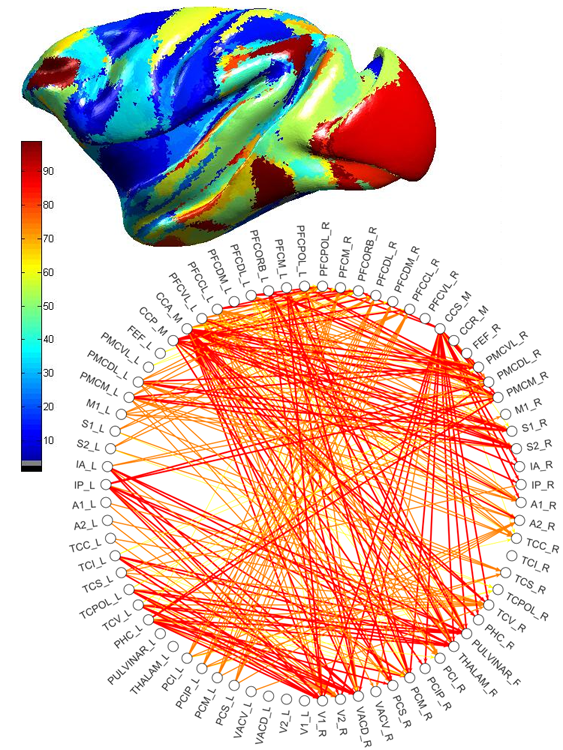
We describe an approach for spatially registering all cortical brain regions from the textual database CoCoMac (Kӧtter, 2004). Currently, it contains 458 studies, mainly tract tracing experiments, many of which define their own cortical (sub)parcellation. Direct registration to a surface-based Macaque cortex is applied to 9 core parcellations using the tool Caret (Van Essen and Dierker, 2007). The rest of the database is semantically linked to these core parcellations using previously developed algebraic and machine learning techniques (Stephan et al., 2000; Bezgin et al., 2008; Bakker et al., ongoing work). For the translation to the human cortex we rely on Van Essen's landmark-based macaque to human warpings. As a result, one can query CoCoMac for a given spatial coordinate in any of the Caret-supported macaque and human cortical templates.
Connectivity was analyzed using multiple graph-theoretical measures to capture global properties of the derived network using the new brain network visualization and analysis tool ConJUNGtion developed on the basis of JUNG software (Madadhain et al., 2005). As a next step, the spatial connectivity maps can be used either as a direct validation of neuroimaging data, or indirectly by constraining generative models of brain activity with a plausible anatomical connectivity assignment.
References
Bezgin G., Wanke E., Krumnack A., Kötter R. (2008) Deducing logical relationships between spatially registered cortical parcellations under conditions of uncertainty. Neural Networks 21(8): 1132-1145.
Kӧtter R. (2004) Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics 2(2), 127-44.
Madadhain J., Fisher D., Smyth P., White S., Boey Y.B. (2005) Analysis and visualization of network data using JUNG. Journal of Statistical Software, Vol. 10, p. 1--35.
Paxinos G., Huang X.-F., Petrides M. and Toga A.W. (2009) The Rhesus Monkey Brain in Stereotaxic Coordinates, 2nd Edition. Amsterdam: Elsevier Science.
Stephan K.E., Zilles K., Kötter R. (2000) Coordinate-independent mapping of structural and functional data by objective relational transformation (ORT). Phil. Trans. R. Soc. Lond. B 355, 37-54.
Van Essen D. C., Dierker D. L. (2007) Surface-based and probabilistic atlases of primate cerebral cortex. Neuron 56, 209–225.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
