From recording to sharing of data - embedding metadata handling into the laboratory workflow using odML
Jan Grewe (Bernstein Center for Computational Neuroscience, Department Biology II, Ludwig-Maximilians-Universität München, Germany), Andrey Sobolev (Department Biology II, Ludwig-Maximilians-Universität München, Germany), Thomas Wachtler (Department Biology II, Ludwig-Maximilians-Universität München, Germany), Jan Benda (Bernstein Center for Computational Neuroscience, Department Biology II, Ludwig-Maximilians-Universität München, Germany)
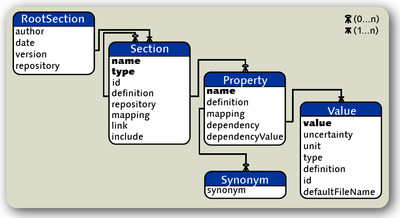
Metadata, such as the type of cell recorded or the stimulus used to evoke a neuronal response, provide essential information about experimental datasets. This information is necessary for the analysis, management and sharing of data. Recently we developed the "Open metaData Markup Language" (odML) aiming at the automated handling of metadata. The odML data model (figure 1) is relatively simple and enables organizing metadata in a hierarchical tree-like structure of "Sections" and "Properties". Properties are extended name-value pairs that allow any information to be included.
Here we will demonstrate how metadata handling can be embedded into the laboratory workflow from the recording of data (using Relacs, www.relacs.net), to data management (LabLog, www.lablog.sourceforge.net) and finally data sharing using the G-Node data management platform (www.g-node.org/data). On our project homepage (www.g-node.org/odml) we provide libraries to read, write and work with odML files in C/C++, Python, and Java (easily used in Matlab) in order to embed metadata handling directly into the tools used in the lab.
The odML defines the format, not the content, so that it is inherently extensible and can be adapted to the specific requirements of any laboratory. For data sharing a correct understanding of metadata and data is only possible if the same terminology is used or if mappings between terminologies are provided. For this purpose we assembled terminologies with definitions of commonly used terms (i.e. Properties). The terminologies can be found on www.g-node.org/odml. Extension and validation of the terminologies can only be achieved in a community approach and we invite everyone for discussion.
To completely describe the conditions that led to a certain dataset requires a vast number of metadata and is thus considered a tedious business. We propose that metadata acquisition should start as early and, as the actual data acquisition, should be as automated as possible, collecting meta information where it becomes available. All our tools and libraries are open source and community feedback is very much appreciated.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
