Knife-Edge Scanning Microscope Brain Atlas: A Web-Based, Light-Weight 3D Mouse Brain Atlas
Chul Sung (Texas A&M University), Ji Ryang Chung (Texas A&M University), David Mayerich (University of Illinois, Urbana-Champaign), Jaerock Kwon (Kettering University), Daniel Miller (Texas A&M University), Todd Huffman (3Scan), John Keyser (Texas A&M University), Louise Abbott (Texas A&M University), Yoonsuck Choe (Texas A&M University)
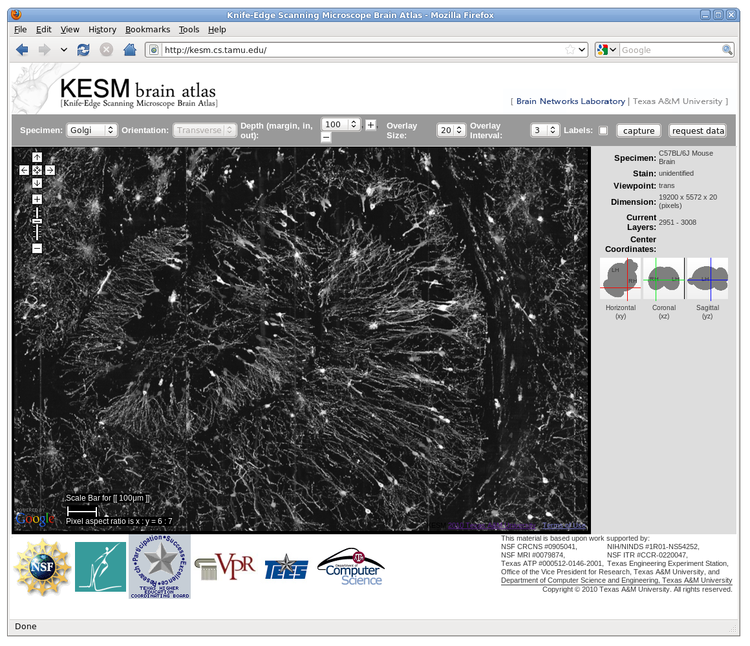
High-throughput, high-resolution 3D microscopy techniques such as the Knife-Edge Scanning Microscopy (KESM) are enabling the acquisition of whole-brain-scale data from small animals such as the mouse. Using the fully automated KESM, we have been able to obtain whole mouse brain data sets at a resolution of 0.6 um x 0.7 um x 1.0 um, enough to show detailed neuronal morphology across the entire brain. Staining for our data sets were done with Golgi (neuronal morphology), Nissl (soma distribution), and India ink (vascular network). In this abstract, we describe a novel web-based 3D atlas we developed to enable broader access to our KESM data sets. The atlas framework, named the Knife-Edge Scanning Microscope Brain Atlas (KESMBA), customizes and extends the Google Maps API to serve the nearly 2TB (per brain) volume data in a multiscale manner, combined with a novel 3D rendering method using image overlays. The standard Google Maps navigation features (pan, zoom, etc.) are supported by default, allowing a multiscale exploration. Individual images in the data sets represent 1um-thin sections, so by themselves, they are not informative. We extended the Google Maps API to overcome this limitation, by overlaying images so that a local 3D volume can be visualized within the browser (see the figure). Other features include scale bar, navigation in the z (depth) direction, overlay control (number of overlays and interval), and location indicator. Experimental features include 3 standard sectioning planes (to be fully supported in the near future). KESMBA requires minimal client-side computing capability (a generic browser with javascript support), thus it enables broader dissemination of the KESM data sets to the neuroscience community. Also, we expect our framework to be general enough to accommodate other data modalities that have large volumes and fine details. On-going and future works include registration of the KESM data sets to the Waxholm space, and custom download and annotation functions.
Acknowledgments: NSF CRCNS #0905041, NIH/NINDS \#1R01-NS54252, NSF MRI \#0079874, NSF ITR \#CCR-0220047, Texas ATP\#000512-0146-2001, Texas A\&M Research Foundation, Texas Engineering Experiment Station, and 3Scan. The appearance of the Google logo in the figure is due to the use of the Google Maps API by the KESM Brain Atlas. Besides that this work was done independent of Google.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
