NeuroMorphoNaut: An Open Source Tool to Make Anatomy Less Tedious
Andrew J. Worth (Neuromorphometrics, Inc.), Gregory L. Millington (Neuromorphometrics, Inc.), Jason Tourville (Boston University)
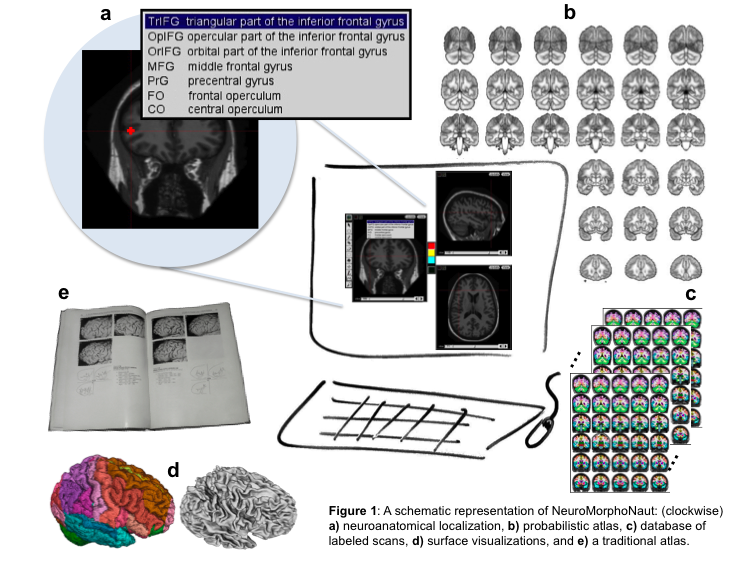
Human brain atlases have evolved from a 2D, physical, single-brain set of pictures and drawings (a book) into something that is digital, stereotaxic, multi-dimensional, has explicitly defined borders, multiple brains, explicitly represents regions and/or surfaces, is searchable, expandable, and has interactive visualization and manipulation capabilities. The problem with current atlases is that they are based on too few scans, define too few regions of interest, and lack a standard labeling protocol. Our solution is "NeuroMorphoNaut" (NMN, see http://neuromorphonaut.org/), a platform for neuroanatomical analysis that consists of 1) a database of manually labeled brain scans, and 2) processing and visualization capabilities. With funding from the NIH, we are manually labeling on the order of 1000 MRI brain scans. Such a large number of scans will establish a quantitative baseline for normative structural measurements against which individual scans can be evaluated. It will enable the creation of "neuroanatomical signatures" for diagnosis and measuring response to treatment in neurology and psychiatry, and also for more general understanding of the correlation of structure with function, age, gender and behavior. Moreover, an understanding of neuroanatomy and accurate anatomical localization are essential in functional imaging studies. This was highlighted by, "In Praise of Tedious Anatomy" (Devlin and Poldrack 2007). Our goal is to deliver open source software that will help address these issues.
To create the database of manually labeled scans, closed borders around regions of interest (ROIs) were created by using isointensity contours and editing. MRI brain scans were obtained from the Open Access Series of Imaging Studies (OASIS). Labeling was performed using a protocol that precisely defines the landmarks, borders and methods for delineating ROIs the entire brain. The probabilistic atlas (see Figure, c) was created using 3D thin plate spline warping with landmarks (AC, PC, midline, and brain extents) to map locations in a scan to corresponding locations in the atlas. The individual structures are related to the atlas as a whole by storing the location of their upper left corners with respect to the landmarks in atlas space.
A cortical parcellation protocol was established (see http://www.braincolor.org/protocols/). Forty scans (20 subjects scanned twice) were manually and comprehensively labeled. A statistical analysis demonstrated the reproducibility of the results. The probabilistic atlas was created using 20 scans. The atlas was used in NMN to create ROIs after warping into the native space of the individual scan. The 20 subjects were used to improve the parcellation protocol by examining the lateral orbital sulcus (los). The location of the posterior end varies with respect to the anterior horizontal ramus of the lateral sulcus (ahls). When los lies medial to ahls, a conflict arises regarding the lateral extent of the inferior frontal region. For the brains examined, the los terminated lateral to ahls in 65% and dorsal 75% in the left hemisphere and 55% in the right.
NeuroMorphoNaut is a modern brain atlas that will enable a deep understanding of normal structure and its variation. It will help address the localization problem in functional neuroimaging by delivering neuroanatomical expertise.
Devlin, J. T. and R. A. Poldrack (2007). "In praise of tedious anatomy." Neuroimage 37(4): 1033-41; discussion 1050-8.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
