Partial Least Squares-Correspondence Analysis (PLS-CA): A New Method to Analyze Common Patterns in Measures of Cognition and Genetics
Derek Beaton (The University of Texas at Dallas), Hervé Abdi (The University of Texas at Dallas)
With
the introduction of large scale studies (such as the Alzheimer's Disease
Neuroimaging Initiative; ADNI) researchers in neuroscience, and genetics are now routinely collecting, and
sharing, a wide variety of very large data sets (e.g., neuroimaging, genomes,
cognitive measures) whose analysis requires suitable statistical methods. In
general, the structure and variety of these data sets preclude the use of
standard hypothesis driven or multivariate techniques (such as multiple
regression or discriminant analysis).
Interestingly,
most of the data in genetics-based cognition research---such as questionnaires
and single nucleotide polymorphisms (SNPs)---are nominal variables and
structured in blocks or tables (e.g., memory tasks for behavior; chromosomes
for a genome). Nominal variables can be analyzed with multivariate techniques
such as Correspondence Analysis (CA) and Discriminant Correspondence Analysis
(DiCA). CA-based methods are to qualitative data what principal components-based
methods are to quantitative data.
However,
CA and DiCA are suitable to analyze one data set. In order to reveal common
information between two data sets (e.g., cognitive measures and genetics) we
adapted the well-known Partial Least Squares Correlation (PLSC) method (suitable
for quantitative measurements) so that it can be used with nominal data. This
new approach, called PLS-CA, integrates in a common framework PLSC and CA.
PLS-CA reveals patterns of co-occurrence of SNPs and behavioral measures --- or
connectivity between genetics and behavior. Furthermore, we extend the PLS-CA
framework to predict group membership (e.g., clinical group). This new
technique, called Discriminant PLS-CA reveals factors common to two nominal
data tables, while accounting for a priori discriminant information for
observations. Discriminant PLS-CA relates causal data (i.e., clinical groups),
to relational data (i.e., genetics and cognition), similar to Multi-block PLSC.
PLS-CA
and Discriminant PLS-CA include inferential non-parametric cross validation
techniques such as permutation tests, jackknifing, and bootstrapping.
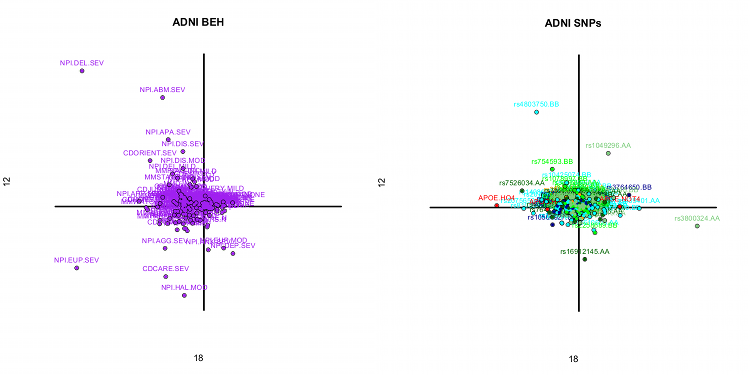
We
illustrate the PLS-CA framework (and its discriminant extension) with data from
the ADNI clinical and genetic datasets. Our goal is to relate genomic (SNPs)
and behavioral data. Results show patterns of genetic variations associated to
both healthy and dysfunctional cognitive traits, as well as genetic variations
for clinical groups.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
