Prediction of Blood Brain Barrier Permeation based on Molecular Indices
Lanting Li (Chinese Academy of Sciences)
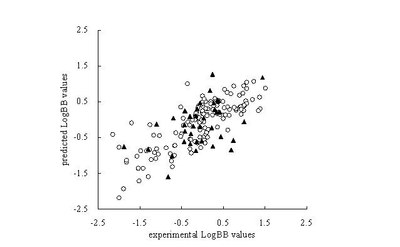
A quantitative structure-activity relationship (QSAR) model for predicting the blood brain barrier permeability in a large variety of compounds (190 compounds) has been developed using molecular indices as predictors. The poor ADME properties are one of the major reasons for the failure of drug candidates in the pre-clinical or clinical phases. The blood brain barrier permeation is one of the important properties for the drugs aimed at central nervous system (CNS) targets. This barrier must be crossed for a therapeutic effect. Conversely, for non-CNS targets, passage across the blood brain barrier (BBB) could lead to undesirable side effects and should be minimized. So a compound’s ability to cross this is of concern to drug designers. To quantitatively predict the BBB permeation and determine the key descriptors largely influencing the BBB permeation in structure, a dataset composed of 190 diversified compounds, was studied by principal component analysis and multivariate linear regression analysis methods. 16 molecular indices which covering 91.59% of all information of descriptors are reached by principal component method, to provide precise and simple guide for the further drug synthesis, 10 descriptors of 16 are selected to build the model with multivariate linear regression method. The resulting model showed satisfactory statistical results with 10 descriptors, and proper predictability were validated by the test set which are showed in figure below. Those key descriptors were identified, and they are valuable and helpful for aiding further screening and development of drugs.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
