The Computable Brain: an interactive website to allow exploration and sharing of functional imaging data
David Gutman (Emory University), Orion Keifer (Emory University), Erin Hecht (Emory University), Joel Saltz (Emory University)
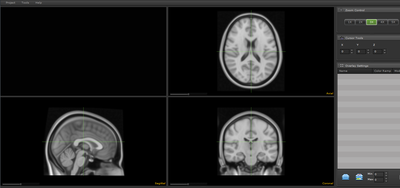
We have developed a prototype application called The Computable Brain that allows visualization and sharing of functional and structural neuroimaging datasets. This application aims to facilitate collaborative data exploration between researchers using a convenient web based GUI. During the data exploration phase of neuroimaging research, a common scenario involves passing "screenshots" of various statistically derived results overlaid on top of an underlying structural image, or in a common reference space. These images may be subsequently marked up using powerpoint or a graphics editor to highlight significant features, and then sent to collaborators for comment. This process is both time consuming and inflexible, since these images are static. Inspired largely by FSLview, we have developed an interactive GUI that allows visualization and sharing of imaging data, as well as a database framework to store files. This obviates the need for collaborators to keep track of multiple files, and more importantly removes barriers associated with the installation of viewing applications, which often require linux experience.
As a prototype, we have loaded several anatomic reference data sets from both human, mouse, and primate species. These images are browseable at http://computablebrain.com. For the MNI152 data set, we are in the process of adding the ability to dynamically query several pre-defined atlas spaces. We have also added the ability to upload both structural underlays as well as functional overlays. The overlays can be dynamically rendered at multiple orientations and thresholds (i.e. the researcher can view the same image at multiple thresholds or orientations by simply updating a slider).
One of the driving applications for this technology centers around the ability to dynamically view and generate probabilistic tractography data generated in the FSL Probtrack software package. The typical workflow in FSL requires a mask or seed region to be drawn, saved, a probabilistic map computed (which can take minutes or hours depending on the ROI size), and then the results redisplayed. Slight changes in the seed region to explore how small changes in ROI placement is relatively labor-intensive, so we have adapted this interface to allow dynamic ROI generation and image rendering in real time. By precomputing the tractography map from every voxel within the brain (~260,000 individual 3-D volumes were stored and processed), ROI’s can be quickly changed and the results immediately displayed. To achieve the dynamic rendering, we use a combination of MYSQL, PHP, javascript, FLEX, and the imagemagick toolkit to allow dynamic image generation and display. Future iterations of this workflow will also allow dynamic computations, so that two (or more) ROI’s can be dynamically rendered and areas showing common and differential connectivity between the ROI’s can be viewed.
Conclusion:
While this resource is still developing, we believe that the concept of social-networking with a low barrier of entry (i.e. all web based) may further facilitate interdisciplaincay collaboration and interpretation of the burgeoning number of neuroimaging result sets that are being generated.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
