The Neuroscience Information Framework (NIF): A Unified Semantic Framework and Portal for Discovery and Integration of Biomedical Data and Resources on the Web
Jeff Grethe (UCSD), Amarnath Gupta (UCSD), Anita Bandrowski (UCSD), Fahim Imam (UCSD), Jonathan Cachat (UCSD), Gordon M Shepherd (Yale), Perry L Miller (Yale), Luis Marenco (Yale), Giorgio A Ascoli (Krasnow Inst. for Advanced Study, George Mason Univ), Paul Sternberg (Caltech), Hans Mueller (Caltech), Arun Rangarajan (Caltech), Maryann Martone (UCSD)
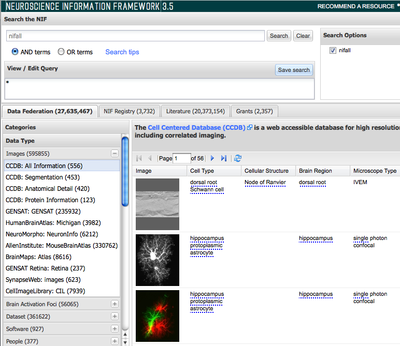
The Neuroscience Information Framework (NIF; http://neuinfo.org) was launched in 2008 to address the problem of finding and integrating neuroscience-relevant resources through the establishment of a semantically enhanced framework to promote resource discoverability and integration. The NIF discovery portal provides simultaneous search across multiple types of information sources to connect neuroscientists and biomedical researchers to available resources. These sources include the: (1) NIF Registry: A human-curated registry of neuroscience-relevant resources annotated with the NIF vocabulary; (2) NIF Literature: A full text indexed corpus derived from the PubMed Open Access subset as well as an entire index of PubMed; (3) NIF Database Federation: A federation of independent databases that enables discovery and access to public research data, contained in databases and structured web resources (e.g. queryable web services) that are sometimes referred to as the deep or hidden web, that are annotated and integrated with a unified system of biomedical terminology. Over the past year, NIF has continued to grow significantly in content, providing access to over 3800 resources through the Registry, and more than 70 independent databases in the data federation, making NIF one of the largest sources of neuroscience resources on the web.
Search and annotation of resources and resource content is enhanced through the utilization of a comprehensive ontology (NIFSTD; http://purl.org/nif/ontology/nif.owl). NIFSTD is expressed in OWL-DL and covers major domains in neuroscience, including diseases, brain anatomy, cell types, subcellular anatomy, small molecules, techniques and resource descriptors. The NIFSTD ontologies are used to refine or expand queries by utilization of the relationships encoded in the ontology. They are served in Ontoquest, a database customized for storing and serving ontologies encoded in OWL. To enable broad community contribution to NIFSTD, NeuroLex (http://neurolex.org) is available as a wiki that provides an easy entry point for the community. The NIF uses a combination of strategies for employing the vocabularies in search and annotation. For the NIF Registry, we use the NIFSTD as a controlled vocabulary for annotating the type and general content of catalog entries. For the NIF databases, we use the ontology to map individual table names, field and values to ontology entities in order to unify search across independent resources that each use their own terminology. NIF also uses the ontology to enhance search through automated expansion of NIF terms using synonyms and abbreviations. The NIF search interface autocompletes search strings with concepts in the NIF ontologies. Through the advanced search interface, users may add additional classes such as parents, children and other related classes. In 2.5, we have incorporated more automated expansions to provide concept-based searches over certain classes of neuroscience concepts. For example, concepts like GABAergic neuron or Drug of Abuse are automatically expanded to include children of these classes, i.e., types of GABAergic neurons. This expansion is done through the provision of logical restrictions within the NIFSTD. When such logical restrictions are present, Ontoquest materializes the inferred hierarchy and auto expands the query to include these classes.
The latest release of NIF provides the foundation for enhanced semantic services. For example, entities contained within the NIFSTD are highlighted within the NIF search results display. Right clicking on these entities will show their semantic category, e.g., organism, anatomical structures, bring up a set of search options. For select entities, e.g. anatomical regions and cells, we have introduced the NIF Cards. NIF cards are specialized search applets that draw upon NIFSTD and the NIF data federation to display additional information about an entity and provide customized search options depending upon the domain. For examples, NIF Cards for anatomical structures will display a definition of the structure, cells contained within that structure and provide informational links to select resources like Brain Info that provide additional information about the brain region. As the NIF Cards evolve, they will provide the basis for linking NIF results into the large ecosystem of linked data.
As the scope and depth of the NIF data federation grows, NIF has been working to create more unified views across multiple databases that export similar information. For example, NIF has registered several databases that provide mapping of gene expression to brain regions or provide connectivity information among brain structures. For these sources, NIF standardizes the database view based on common data elements and column headers. In performing this type of integration across resources, NIF has had to confront the different terminologies utilized by different resources. While we don’t change the content of the database, we do provide some mappings to the NIF annotation standards for recognized entities such as brain regions and cell types. We have also started to provide a set of standards for annotating quantitative values such as age and gene expression level. Such translation is necessary to allow users to search for qualitative terms such as “Adult” and “Increased gene expression” through the query interface but return results from databases that report only the age of the organism or some sort of measure for gene expression. To do this, NIF has defined age ranges for adulthood in common laboratory species such as the mouse and a standard set of categories for expression levels.
In this presentation, we will demonstrate the NIF discovery interface and some of its registration tools. Resource providers are encouraged to register their resources to NIF using some of its custom registration tools and to view and comment on the NIF annotation standards.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
