Validation of Granger Causality within In Vitro Engineered Feed-Forward Cortical Networks
Sankaraleengam Alagapan (University of Florida), Liangbin Pan (University of Florida), Bruce Wheeler (University of Florida), Thomas DeMarse (University of Florida)
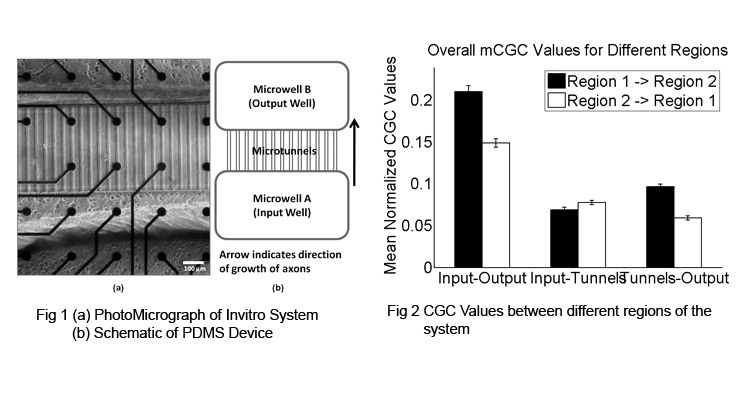
Understanding the functional connections between different regions of the brain presents a major challenge in neuroscience. A number of statistical techniques have been developed to determine functional connectivity among populations of neurons. However validation of these techniques has typically relied on either macroscopic anatomical information or computer simulations for want of a living system where connectivity is known or constrained a priori. Fabrication of special devices [1] in our lab has enabled us develop dissociated cultures of neurons which are predominantly unidirectional, and when coupled with 60 electrode microelectrode arrays (MEA), the extracellular recordings from this in vitro system (Fig 1) provide a valuable platform to validate these statistical measures. These devices, fabricated from PDMS, are composed of two microwells, each containing approximately 20,000 neurons, connected by 51 microtunnels with dimensions: 3 x 10 x 400 µm (h x w x l). The devices were then placed over an 8x8 grid of electrodes (Multichannel Systems, electrode spacing 200 um) in which electrodes could simultaneously measure spike propagation along the tunnels and activity within and between each well. To create unidirectionally connected cortical populations, one well (input) was plated with E-18 cortical neurons and after axons had extended into the tunnels at 10 days in vitro, the second well (output) was plated. In essence, this creates a simple feedforward network in which the number of connections can be manipulated by the number of tunnels and direction determined by the time at which neurons are plated. Extracellular signals (spikes) were recorded using the MEA. Network activity consisted of both tonic spikes and synchronized bursts. Each spike train was smoothed with an exponentially decaying signal to obtain a continuous waveform. Conditional Granger Causality (CGC) between different electrodes was then computed using the GCCA toolbox [2] and results compared to conventional cross correlogram analysis. CGC estimates of causal strength within tunnels and between input and output wells were higher in the predicted feed-forward direction (Fig 2) and paralleled results produced by conventional cross-correlation metrics.
References:
[1] B.J. Dworak and B.C. Wheeler, “Novel MEA platform with PDMS microtunnels enables the detection of action potential propagation from isolated axons in culture,” Lab on a Chip, vol. 9, 2009, pp. 404-410.
[2] A.K. Seth, “A MATLAB toolbox for Granger causal connectivity analysis.,” Journal of neuroscience methods, vol. 186, Feb. 2010, pp. 262-73.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
