Visual Informatics and Computational Genomics using the Graphical Pipeline Environment
Ivo Dinov (UCLA), Federica Torri (UCI), Fabio Macciardi (UCI), Alen Zamanyan (UCLA), Arthur Toga (UCLA)
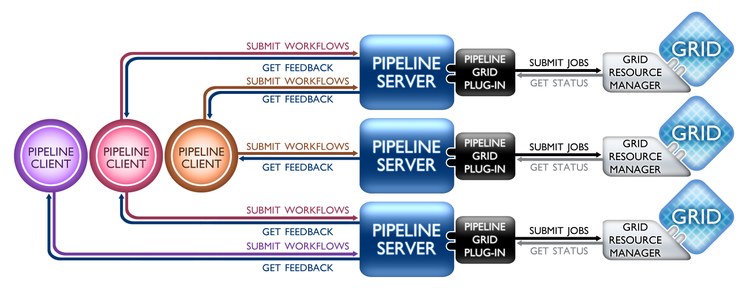
Contemporary informatics and genomics research require efficient, flexible and robust management of large heterogeneous data, advanced computational tools, powerful visualization, reliable hardware infrastructure, interoperability of computational resources, and detailed protocol provenance. The Pipeline Environment (http://Pipeline.loni.ucla.edu) is a client-server distributed computational environment that facilitates the visual graphical construction, execution, monitoring, validation and dissemination of advanced data analysis protocols. This presentation will demonstrate hands-on several informatics and genomics applications via the Pipeline environment and emphasize the graphical management of diverse genomics tools, the interoperability of informatics tools, and Grid resource management. Examples of tools that will be showcased include EMBOSS, mrFAST, GWASS, PLINK, MAQ, SAMtools, Bowtie, GATK and others (http://pipeline.loni.ucla.edu/support/pipeline-workflows/). The Pipeline Environment may be tried out directly online without any account settings or software installation requirements (http://Pipeline.loni.ucla.edu/PWS).

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
