Visualization and exploration of the transcriptomic landscape of the human brain
Chris Lau (Allen Institute for Brain Science, WA), Tim Dolbeare (Allen Institute for Brain Science, WA), Felix Lee (Allen Institute for Brain Science, WA), David Feng (Allen Institute for Brain Science, WA), Tim Fliss (Allen Institute for Brain Science, WA), Changkyu Lee (Allen Institute for Brain Science, WA), Angela Guillozet-Bongaarts (Allen Institute for Brain Science, WA), Zackery Riley (Allen Institute for Brain Science, WA), Barry Daly (University of Maryland School of Medicine, Baltimore, MD), Rao Gullipalli (University of Maryland School of Medicine, Baltimore, MD), Alan McMillan (University of Maryland School of Medicine, Baltimore, MD), Elaine Shen (Allen Institute for Brain Science, WA), Mike Hawrylycz (Allen Institute for Brain Science, WA), Chinh Dang (Allen Institute for Brain Science, WA), Allan Jones (Allen Institute for Brain Science, WA), Lydia Ng (Allen Institute for Brain Science, WA)
Introduction
The Allen Human Brain Atlas is an online multimodal atlas of the human brain that integrates anatomic and genomic information. A key component of the resource is a comprehensive dataset of microarray-based gene expression profiles from two non-diseased postmortem brains resulting in over 100 million data points. The visualization challenge of presenting this dataset online is to effectively allow users to explore the transcriptomic landscape of the brain in connection to its 3D anatomy and supporting histological image information.
Methods
Two postmortem brains were imaged using a 3T Siemens Trio MR scanner. T1, T2, and diffusion tensor images were acquired. The tissue was then frozen and cut into approximately 1cm coronal slabs. Each slab was further sub-divided into 2”x3” blocks. The blocks were sliced at 25mm thickness and the tissue Nissl stained and scanned at 1 µm/pixel on an Aperio ScanScope XT microscope. Images were annotated to delineate sampling areas for microarray analysis as well as to provide anatomic information for online presentation. For each block, expert annotators provided 10-50 corresponding landmark locations between the 2D block-based image and the 3D MRI. These landmarks were used to estimate deformation fields using thin-plate splines. Thus, any position on any block could be mapped back to the 3D anatomical context. RNA isolated from each sampling area (~1000 per brain) was hybridized to a custom microarray chip to measure gene expression over the whole transcriptome.
Results
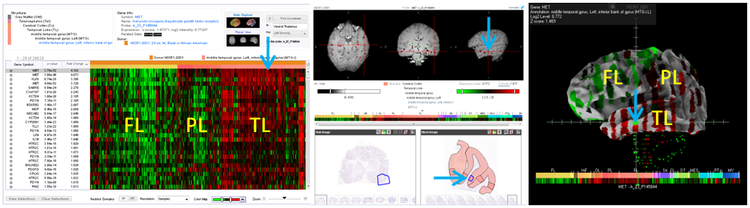
A web application has been implemented in which expression data is visualized as color-coded heatmap matrices. Each row represents a probe from the array and each column represents a sample or structure. The color in each cell of the matrix represents expression level either in log2 scale or as z-scores per probe. A novel heatmap service engine was implemented to generate tiles of the heatmap as needed by the web application. When a user initiates a query on the application, a heatmap session is established. As the user scrolls through the results, tiles of the heatmap are served much like web-based street map applications. Three types of queries are currently supported: (1) search by gene name or category, (2) search by differential enrichment expression of one region compared to another region and (3) correlative search against a seed probe to find probes with a similar expression profile.
Associated image data can be browsed in the multiplanar viewer. The top panel is an interactive cardinal plane view of the MR data with the centroid of sampling areas overlaid and color-coded by the expression value of the selected probe. Clicking on a sample point in the MRI displays the associated high resolution histological data in the 2D image viewer in the lower panel. The corresponding sample area in the image is highlighted, allowing the user to make the connection between cytoarchitecture and gene expression. Expression profiles and brain anatomy can also be viewed in 3D via Brain Explorer®. To aid visualization of gene expression patterns in the cortex, expression is rendered on a partially inflated white-matter surface.
All data in the Atlas can be freely accessed at http://human.brain-map.org. More data will be added during this 5-year project with the goal of incorporating an additional eight brains as well as continued development of visualization and search tools.

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
