Web-based collaborative neuronal reconstruction with CATMAID
Stephan Gerhard (Institute of Neuroinformatics, UZH / ETHZ, Zürich), Mark Longair (Institute of Neuroinformatics, UZH / ETHZ, Zürich), Stephan Saalfeld (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden), Pavel Tomancak (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden), Albert Cardona (Institute of Neuroinformatics, UZH / ETHZ, Zürich)
Reconstructing
neuronal circuits at such high resolutions that synaptic connections
are clearly visible can currently only be done from image data acquired
via electron microscopy (EM). These stacks of images enable precise 3D
reconstructions of neuronal morphology. While automatic methods for
segmenting such images are certainly improving, much annotation and
segmentation still needs to be done by human operators carefully
examining the images. In addition, the EM data sets that must be dealt
with are often many terabytes in size. The requirement for hundreds of
annotators to each have a local copy would be prohibitively expensive.
To address these requirements, we have extended CATMAID, the
Collaborative Annotation Toolkit for Massive Amounts of Image Data,¹ to
allow many researchers to trace neurons collaboratively in the same data
set.
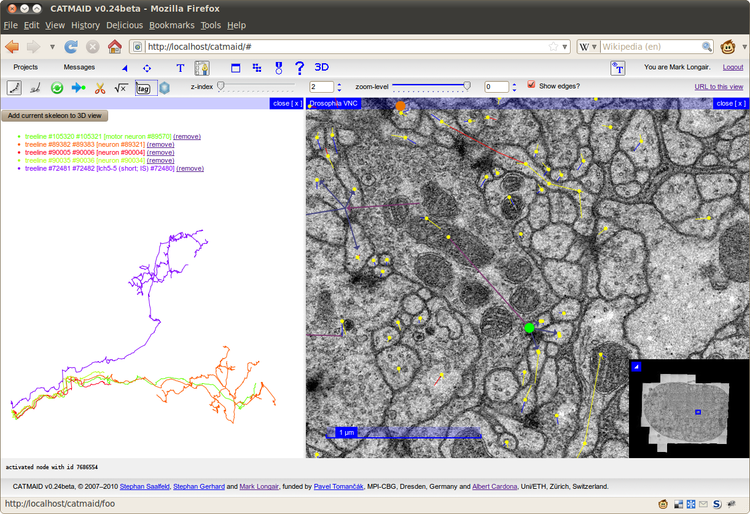
CATMAID
is a web-based system, so each annotator only needs a web browser and
minimal local storage space requirements. The system already provides an
elegant Google Maps-style interface for browsing huge image stacks,
collaborative text annotation and a simple interface for asynchronous
server-side jobs. We have added two further types of annotation
primitive: skeletons and connectors. Skeletons are tree structures
suitable for representing the midlines of neuronal arborizations.
Connectors link the nodes of skeletons in a many-to-many relationship
through a central point, and are suitable for representing polyadic
synapses. These annotations are stored in a logical hierarchy, which
can be arranged by the researchers in the web interface in order to best
represent the structure and biological understanding of the tissue
under examination.
Every
new or changed annotation is immediately reflected in the remote
centralized database, so that each researcher always sees up-to-date
annotations. Hundreds of annotators can thus concurrently reconstruct
the many neurons that make up a circuit. The centralized storage of
annotations also allows automatic incremental backups and making the
data available via web services.
We
also have added in-browser 3D visualization of the skeletons,
text-tagging of skeleton nodes and connectors, and statistics showing
the progress of the tracing. Skeletons can be exported in the standard
SWC format for further analysis.
While
we have tailored the user interface of CATMAID for tracing the midlines
of neurons and adding synapses, the annotation primitives of skeletons
and connectors are stored in the database as elements of tissue-agnostic
subject-predicate-object relations, which means both that the system
can be easily adapted to different annotation tasks, and that the data
can be made accessible via semantic web technologies.
We
present, as an example, our progress in tracing a terabyte serial
section TEM (Transmission Electron Microscopy) data set from one
abdominal segment of the ventral nerve cord of a first instar Drosophila larva.
¹ Saalfeld S, Cardona A, Hartenstein V, Tomancák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25: 1984–1986

 Latest news for Neuroinformatics 2011
Latest news for Neuroinformatics 2011 Follow INCF on Twitter
Follow INCF on Twitter
